Challenge 09: Dynamic programming
Inspiration
As discussed in class, sequence alignment is arguably the most important and most used algorithm in bioinformatics. The challenge below is inspired from one part of the 2nd homework from COSC494/594 being offered again this spring.
Note sequence alignment is very similar to common interview questions on strings including but not limited to edit distance (class), longest increasing subsequence, and longest common subsequence.
Requirements
You should download the two sequence pairs (4 sequences total) from Github.
We will be performing global alignment with +1 for a match, -1 for a mismatch, and -1 for any gap.
Your goal is to compute the value of the
(m, n) cell using dynamic programming and the base case/recurrance
discussed in class. Please review these notes for the global alignment recurrance and
some examples. Additional info is also on Wikipedia here
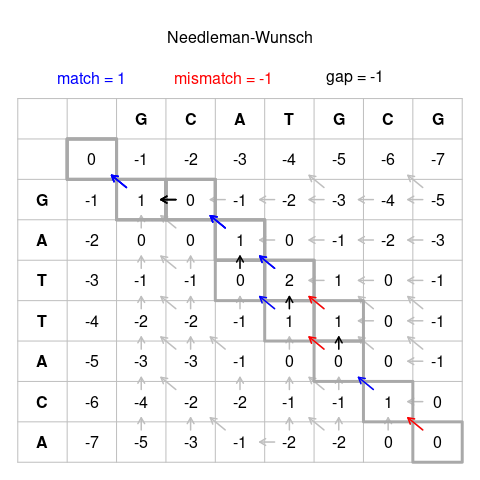 Source Wikipedia
Source Wikipedia
Unlike 494/594 you will not be required to produce a traceback/actual alignment as shown above but please come and see us if you are curious and would like to do it for fun.
Rubric
We will grade this challenge using the rubric below.
+2 Code is commented +2 initialize matrix properly +4 computes diagonal, other gap cases correctly +6 returns correct score for simple alignment (make test) +6 returns correct score for second pair of real sequences (seq1/seq2)
To aid in the grading process please include a makefile in your submission that compiles your code and provide a brief README on how to run your code on the two real sequences and what results to expect.
Submission
To submit your solution, you must submit a single archive (.tar, .gz) file
on Canvas prior to the deadline with the required README included.